PROVIDENCE, R.I. [Brown University] — Gene expression, DNA repair, and protein making are intricate and vital processes, so it’s a bit of a surprise in a new Brown University study that those processes depend significantly upon proteins that do not take a static form. Instead, FUS — Fused in Sarcoma’s — protein interaction region remains forever squirmy and squiggly, the study reports, performing its healthy functions by assembling into liquid-like droplets with other copies of itself and key molecules such as RNA.
The findings, published in the journal Molecular Cell, may dash hopes that preventing FUS from causing ALS and cancer could be a typical drug development task of resolving its errant forms and then figuring out how to block them. Instead, biologists may have to understand and control variations, or codes, of amino acid sequences in the protein that apparently guide them to engage in certain kinds of assemblies and prevent droplet assemblies from turning into tangled knots.
“The cell, we know, organizes these structures transiently,” said study senior author Nicolas Fawzi, assistant professor of medical science in the Department of Molecular Pharmacology, Physiology, and Biotechnology at Brown University. “That code is malleable. If we could understand what holds these things together, we might be able to change the code on the fly the way the cell does.”
Huddling up
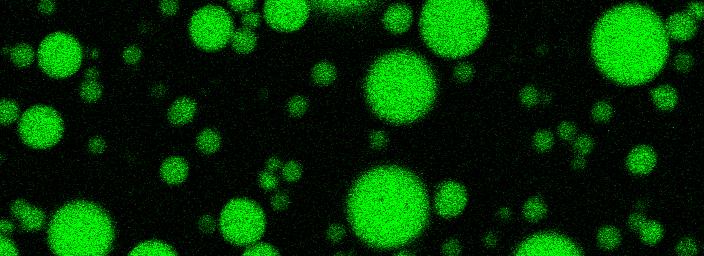
Assemblies of FUS strands, which appear under the microscope to be little droplets, rapidly form and dissipate throughout cells as needed to bind and organize RNA within the cell. For instance, one kind of assembly may form to shepherd mRNA from the nucleus to the ribosome. Another may form to capture RNA and sequester it for a time, Fawzi said. Individual FUS proteins, which are like little noodles, are found all over the place, floating in wait for the moment when they become cued into action.
“The formation of these droplets of proteins and RNA together somehow seems to be important for getting RNA through its lifecycle, getting it made, getting it where it needs to be, and getting it transported and processed properly,” Fawzi said.
Before this study, scientists knew the protein formed these little droplets to organize RNA and knew that sometimes FUS can go awry and become associated with disease. In the case of ALS they transform into intractable and accumulating knots. In some cancers, like leukemias and sarcomas, Fawzi said, mutated FUS proteins may form droplets in unfortunate locations, leading to misregulated genetic transcription. The new study shows FUS droplets can draw in RNA polymerase II, a major transcriptional protein.
Droplets of FUS flow, form and fuse in the lab.
What scientists didn’t know until Fawzi’s team looked into the little droplets, however, was that the FUS proteins take no characteristic form, even when they are performing their RNA chaperone functions. Instead, they only produce functional droplet structures when many copies of FUS along with RNA combine into these droplets.
“If a cell membrane is more like a wall, then a droplet is more like a huddle, where everyone takes their two hands and grabs on to someone else’s shoulders in a random array,” Fawzi said. “That would keep it together — the proteins and RNA self-organizing with each other, rather than all being enclosed.”
Fawzi, lead author Kathleen Burke, and their co-authors used both nuclear magnetic resonance spectroscopy, which can define the structure of biomolecules with atomic resolution, and fluorescence microscopy to make their observations about the perpetually unstructured lives of FUS proteins and their droplet-like huddles.
Cracking the code?
The protein’s propensity to huddle into one kind of spherical arrangement vs. another appears to depend on its sequence of amino acids. Small repeating areas of amino acids work to form little strips of attachment along the protein strand. That’s where the proteins can grab each other. Different arrangements of those positions may lead to the different functional formations.
“There is some kind of code there, but we don’t know how to read that code yet,” Fawzi said. “We can’t yet predict what the function of the protein is going to be or who might interact with whom, or which partners might join into droplets together.”
In the lab Fawzi is now studying how altering amino acid sequences affects FUS behaviors in hopes that it will lead to a clinically useful strategy against ALS and cancer. His prime targets are the mutations in FUS that are reported to cause some forms of ALS.
Fawzi said funding and mentoring through two multidisciplinary biomedical research centers based in Rhode Island were integral to allowing the idea of the study to reach completion. The Center for Biomedical Research Excellence in Skeletal Health and Repair and the Rhode Island IDeA Network for Excellence in Biomedical Research are supported by the National Institutes of Health’s Institutional Development Award (IDeA) program under grant numbers P20GM104937 and P20GM103430, respectively. These programs build research capacity in states that historically have had low levels of NIH funding by supporting basic, clinical, and translational research; faculty development; and infrastructure improvements.
“This is an example of how the infrastructure development and collaborative network formation supported by the program can enable exciting and productive team approaches to address complex questions and move biomedical science forward,” said W. Fred Taylor, who directs the IDeA program at NIH’s National Institute of General Medical Sciences.
Additional funding for the study came from the Rhode Island Foundation and Brown University. Some of the work occurred in the Brown University Structural Biology Core Facility, the Rhode Island NSF/EPSCoR Proteomics Shared Resource Facility, and Brown’s Leduc Bioimaging Facility and genomics core facility.
In addition to Fawzi and Burke, the paper’s other authors are Abigail Janke and Christy Rhine.