PROVIDENCE, R.I. [Brown University] — A computerized epidemiological model of the spread of the mosquito-borne West Nile virus in 17 counties of California in 2005 successfully predicted where 81.6 percent of human cases of the disease would arise and defined high-risk areas where the risk of infection turned out to be 39 times higher than in low-risk areas, according to newly published research. The DYCAST software used in those predictions is now open-source and is being applied to other diseases.
“One of the things that really differentiates DYCAST from other approaches is that it’s based on biological parameters,” said Ryan Carney, a Brown University graduate student who is the lead author on a paper about DYCAST’s performance that appears in the current issue of the journal Emerging Infectious Diseases, published by the Centers for Disease Control. “All of the parameters in the model are based on experimental data related to the biology and ecology of the virus, mosquito vector, and bird host.”

For example, the spatial parameters of the model include how far mosquitoes and infected birds are likely to fly. Key time parameters include how long the virus needs to incubate in mosquitoes before they become infectious and the lifespan of infected birds. Carney said that by using biology to define the geographic and temporal attributes of the model rather than county or census tract borders, which are convenient for humans but irrelevant to birds and mosquitoes, the model allowed the California Department of Public Health to provide early warnings to an area stretching from the Bay Area through Sacramento to the Nevada line, as well as regions in southern California.
Carney implemented the software when he worked for the California department in 2005. (The software was created by Constandinos Theophilides at the City University of New York.) Feeding the model in 2005 were 109,358 dead bird reports phoned in or entered by members of the public via a state hotline and website.
As more dead birds were reported in close proximity, the software would generate daily maps of areas at high risk for human infection, providing an early warning to local public health officials. The software, for example, predicted areas as high-risk more than a month before the first human cases arose, on average.
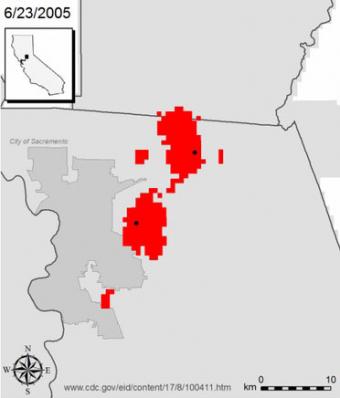
Predicting the course of disease
DYCAST software generated daily maps of high West Nile virus risk areas (red) and human cases (black dots) in the greater Sacramento area in 2005. The animation condenses four and a half months into just over a minute.
In Sacramento County, location of the largest West Nile virus epidemic in the United States that year, DYCAST helped mosquito control officials target their testing and spraying resources — actions that ultimately reduced human illness, Carney said.
After 2005, the department implemented the model throughout the state, although the number of human cases and reported dead birds, along with the model’s prediction rates, dropped sharply.
In 2007 Carney enrolled as a master’s student at Yale and adapted the DYCAST model to track dengue fever in Brazil, using a version of the software that his CUNY collaborators had converted to an open-source platform. With the specific parameters of that disease, DYCAST was able to predict its spread in the city of Riberão Preto in Brazil, Carney said, citing unpublished data.
Carney has continued his analysis and development of DYCAST and dengue at Brown, where he is a doctoral student of ecology and evolutionary biology. He said the software at its core has potential to be adapted as an early warning system for other infectious diseases or even bioterrorism attacks.
In addition to Carney, other authors on the paper include Sean Ahearn and Alan McConchie of CUNY (McConchie is now at the University of British Columbia–Vancouver), Carol Glaser, Cynthia Jean, Kerry Padgett, Erin Parker, Ervic Aquino, and Vicki Kramer of the California Department of Public Health, and Chris Barker and Bborie Park of the University of California–Davis.
The Centers for Disease Control funded the research.