PROVIDENCE, R.I. [Brown University] — Even on the evolutionary time scale of tens of millions of years there is such a thing as being in the right shape at the right time. An anatomical difference in the ability to seize the moment, according to a study led by Brown University biologists, explains why more species in one broad group, or clade, of grasses evolved a more efficient means of photosynthesis than species in another clade.
Their findings appear this week in the Proceedings of the National Academy of Sciences.
Biologists refer to the grasses that have evolved this better means of making their food in warm, sunny, and dry conditions with the designation “C4.” Grasses without that trait are labeled “C3.”
What scientists had already known is that while all of the grasses in the BEP and PACMAD clades have the basic metabolic infrastructure to become C4 grasses, the species that have actually done so are entirely in the PACMAD clade. A four-nation group of scientists wondered why that disparity exists.
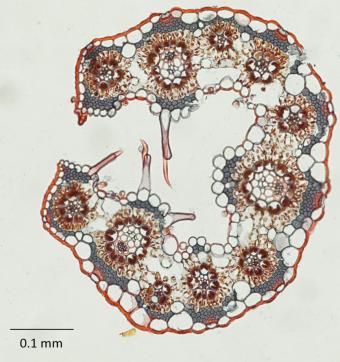
To find out, Brown postdoctoral researcher and lead author Pascal-Antoine Christin spent two years closely examining the cellular anatomy of 157 living species of BEP and PACMAD grasses. Using genetic data, the team also organized the species into their evolutionary tree, which they then used to infer the anatomical traits of ancestral grasses that no longer exist today, a common analytical technique known as ancestral state reconstruction. That allowed them to consider how anatomical differences likely evolved among species over time.
Paradoxically, to understand C4 evolution, the researchers focused on the anatomy of C3 grasses in each clade.
In general what they found was that in the leaves of many PACMAD C3 grasses the veins were closer together, and that the veins themselves were surrounded by larger cells (“bundle sheath” cells) than in BEP C3 grasses. Ultimately PACMAD grasses had a higher ratio of bundle sheath cells to mesophyll cells (cells that fill in the area between veins).

In C4 plants, such an anatomical arrangement facilitates a more efficient transfer and processing of CO2 in the bundle sheath cells when CO2 is in relatively short supply. When temperatures get hot or plants become stressed, they stop taking in as much CO2, creating just such a shortage within the leaf.
So PACMADs as a group had developed an anatomical predisposition to C4 photosynthesis that BEP grasses didn’t, said senior author Erika Edwards, an assistant professor of ecology and evolutionary biology at Brown.
“We found that consistently these PACMAD C3s are very different anatomically than the C3 BEPs,” she said. “We think that was the evolutionary stepping stone to C4-like physiology.”
When the new leaves turned over
It wasn’t always this way. Back around 60 or so million years ago, BEP and PACMAD grasses were more similar and both headed in the same direction. The distance between the leaf veins in both clades had been growing closer together. But then they started to diverge in a key way. The bundle sheath cells surrounding the veins in BEP grasses started to shrink down while those in PACMAD grasses stayed larger.
For a long time the climate didn’t particularly punish or reward either of those directions. But then the climate changed and opportunity knocked, Edwards said. Only PACMAD was near the proverbial door.
“When atmospheric CO2 decreased tens of millions of years after the split of the BEP and PACMAD clades, a combination of shorter [distances between veins] and large [sheath] cells existed only in members of the PACMAD clade, limiting C4 evolution to this lineage,” Christin and co-authors wrote in the paper.
The researchers also found that evolution among C4 grasses was anatomically nuanced. Some C4 grasses evolved because of advantageous changes in outer sheath cells, while others saw the improvement in inner sheath cells.
Ultimately, Edwards said, studies like this one show that plant biologists have made important progress in understanding the big picture of when and where important plant traits evolved. That could lead to further advances in both basic science and perhaps agriculture as well.
“Now that we have this increasingly detailed bird’s-eye view, we can start to become a more predictive science,” she said. “Now we have the raw goods to ask interesting questions about why, for example, one trait evolves 10 times in this region of the tree but never over here. In terms of genetic engineering we’re going to be able to provide some useful information to people who want to improve species, such as important crops.”
In addition to Christin and Edwards, the paper’s other authors at Brown were David Chatelet and Laura Garrison. Other authors were Colin Osborne of the University of Sheffield in the U.K.; J. Travis Columbus of Claremont Graduate University in California; Guillaume Besnard of the Universite Paul Sabatier-Ecole Nationale de Formation Agronomique in Tolouse, France; Trevor Hodkinson of Trinity College in Dublin, Ireland; and Maria Vorontsova of the Royal Botanic Gardens in Surrey, U.K.
The National Science Foundation (grants 0920147 and 0843231), the Marie Curie International Outgoing Fellowship (252568), and the Agence Nationale de la Recherche (10LABX-41) supported the research.