PROVIDENCE, R.I. [Brown University] — The fusion of a sperm cell with an egg cell is the very first step in the process that leads to new individuals in sexually reproducing species. Fundamental as this process may be, scientists are only now beginning to understand the complexities of how it works.
In a paper published in PLOS Biology, researchers have described the detailed structure of proteins that enable sperm-egg fusion in two different species: a flowering plant and a protozoan. The researchers hope that revealing the process in these species and their relatives might bring scientists a step closer to understanding it across sexual species, including humans and other vertebrates.
“It’s surprising to me that we still don’t know how a human sperm fuses with a human egg,” said Mark Johnson, an associate professor of biology at Brown University and a study co-author. “One of the things we hope this paper will do is establish a structural signature for the proteins that make gamete fusion work in these species so that we might be able to look for it in species where those protein mechanisms are still unknown.”
Johnson has been working for years to understand gamete (sperm and egg) fusion. In the early 2000s, he identified a protein on the sperm membrane of the flowering plant species Arabidopsis thaliana that seemed to have some influence on the gamete fusion process in that species. His work showed that a mutation in the gene the produces the protein, known as HAP2, causes sperm become unable to fuse with the Arabidopsis egg.
A few years later, Kristin Beale, a Brown graduate student working with Johnson, predicted that HAP2 was related to the protein that viruses use to bore their way into host cells. That makes sense, Johnson says, since both sperm and viruses need to have a mechanism for inserting themselves into a cellular membrane.
Since those initial discoveries, Johnson and other researchers have searched the genomes of other organisms for gene sequences that look similar to the Arabidopsis HAP2. They’ve found similar sequences in a wide variety of eukaryotes (organisms whose cells have a discrete nucleus) — many plant species, algae, insects and some animals. It was conspicuously absent however in vertebrates, including humans.
It’s possible, Johnson says, that vertebrates do indeed have a HAP2-like protein, but its genetic sequence may have changed so much over evolutionary history that it’s hard for researchers to spot by sequence alone. So rather than searching for genes that produce the protein, it might be better to look at the structure of proteins themselves — to search for vertebrate proteins that are structurally similar to those of the HAP2 proteins that have already been identified. But that requires the structures of known HAP2 proteins to be resolved in detail, which is what Johnson and his colleagues set out to do with this latest study.
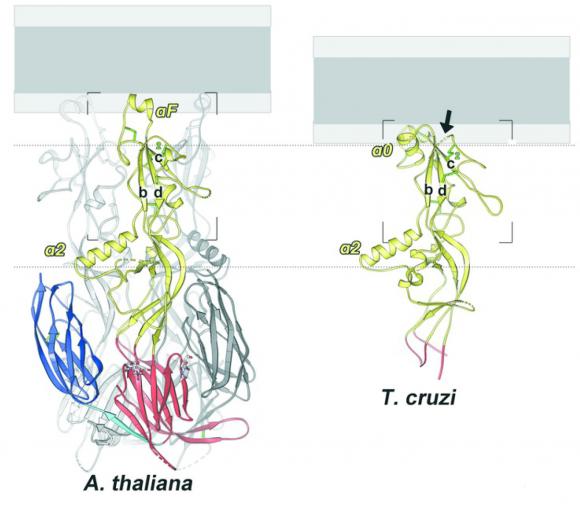
Johnson’s lab worked with the lab of Felix Rey, at Institut Pasteur in Paris, where graduate student Juliette Fedry resolved the structures of HAP2 proteins from two distantly related eukaryote species: Arabidopsis and Trypanosoma cruzi, a protozoan parasite. To resolve the structures, the team in Paris used a technique called x-ray crystallography, which involves crystallizing the proteins and then observing how the crystals scatter x-rays. The structure of the protein can be observed from the scattering pattern. Rey’s lab specializes in the technique, particularly for imaging the viral fusion proteins that are related to HAP2.
Jennifer Forcina, a graduate student in Johnson’s lab, took advantage of the novel structural data to determine how HAP2 drives fertilization in flowering plants, defining the amino acids at the tip of the protein that insert into the egg membrane.
The study found that while the basic structure of the HAP2 proteins from the two species was broadly similar, they had evolved to be different in key areas. Specifically, the tips of the proteins — the parts that are thought to first pierce the membrane of an egg cell — were substantially different. While the flowing plant HAP2 employs a single helical structure at the tip, the protozoan version has three small loops that hook into the target membrane. While the differences in insertion mechanisms between the two proteins shed new light on how HAP2 works, the similarities between the two are important in looking for HAP2 in other organisms, Johnson says.
“At this point we have two kinds of organisms: those for which we know how gamete fusion works — and they all have HAP2 — and organisms for which we don’t know how gamete fusion works, none of which have a recognizable HAP2,” Johnson said. “Now that we know what’s conserved structurally in these two proteins that differ at the sequence level, we can think about looking for those structures to look for related proteins in other species.”
That includes humans, Johnson says. “We just need to get more structures for human proteins so we can look for common structural features.”
Other authors on the paper were Pierre Legrand, Gérard Péhau-Arnaudet, Ahmed Haouz and Thomas Krey.